In this work, we propose BigFiRSt (Big data based Flash and peRf algorithm for mining SSRs), which is a novel Hadoop-based program suite and is specifically designed to integrate paired-end reads merging and SSRs search into an effective computational pipeline.
Intuitively, it is more convenient for biologists to process and analyse large-scale sequences by a user-friendly web interface. However, in practice, it remains a challenging problem for users to upload large scale datasets from their local machines to the online web server.
In order to facilitate users to merge read pairs and subsequently identify SSRs in relatively small datasets, here we provide a publicly available web interface of BigFiRSt. To the best of our knowledge, there is no other such web interface integrating these two processes currently available in the research community.
On the other hand, for handling very large datasets and facilitating the data process using local computers, we also provide the source codes of BigFiRSt for download at https://github.com/JinxiangChenHome/BigFiRSt, such that users can configure and execute the BigFiRSt program on a cluster supported by the Hadoop or a public cloud platforms.
The overall framework of the BigFiRSt methodology is illustrated in Figure 1. BigFiRSt contains two modules: BigFLASH (Figure 1A) and BigPERF (Figure 1B). BigFLASH is used to merge short read pairs generated by NGS platforms and output long consensus reads, while BigPERF extracts SSRs that are located
between the flanking regions of a predefined length from large-scale reads. These modules can be further integrated into a pipeline that takes the output of BigFLASH as the input to BigPERF. The red line in Figure 1 highlights the pipeline that connects BigFLASH with BigPERF.
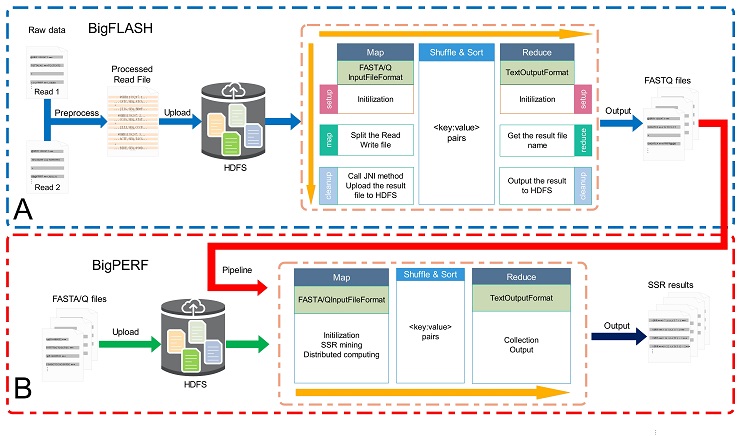 Figure 1. The overall framework of the BigFiRSt methodology.
Figure 1. The overall framework of the BigFiRSt methodology.Here, we show a short tutorial on how to use this web server.
The main page has seven sections (Home, Online Service, Search, Job List, Download, Help and Contact). The details are shown in Figure 2..
 Figure 2. Main interface of the web server.
Figure 2. Main interface of the web server. 1.Home: brief descriptions of the framework of BigFiRSt.
2.Online Service: provides the online operations of the FLASH, PERF, the pipeline.
3.Search: searches the submitted job.
4.Job List: displays all submitted jobs.
5.Download: to download the source code of BigFiRSt.
6.Help: provides flow charts or text informing users how to use the web server.
Contact: methods for users to contact us if they have questions or would like to request more information about this website.
2.1 Online FLASH usage
Please refer to the following steps (Step 1 to 4) Online FLASH usage.
Step 1: Upload two FASTQ files or input sequences in the text area. For FLASH,
users must provide a dataset by uploading two FASTQ files or input two FASTQ sequences. If you don’t know how to do this,
you can just click “example”, and some sample sequence inputs will be showed, as in Figure 3. In the meantime,
users can also choose to upload two FASTQ files, and an illustration of this is shown in Figure 4.
It's worth noting that you need to make sure the input box is empty before uploading the files.
If it is not empty, please click ‘clear’ to clear the previously entered sequence and then upload the files.
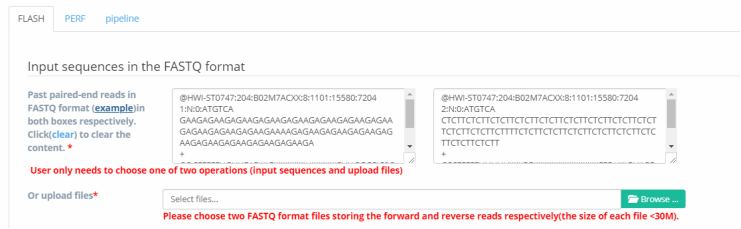 Figure 3. Input read pairs with FASTQ format.
Figure 3. Input read pairs with FASTQ format. Figure 4. Uploading two files in FASTQ format.
Figure 4. Uploading two files in FASTQ format.Step 2: Input values of different kinds of parameters in the designated text area and select descriptors. For convenience, there are many descriptor types to choose from. Text areas should be chosen or filled. For some options with checkboxes, if users want to apply these options, you can just check “YES”. For parameters with text area, users can just skip when they have no need to use them (that will be interpreted as a null value). The details are as shown in Figure 5.
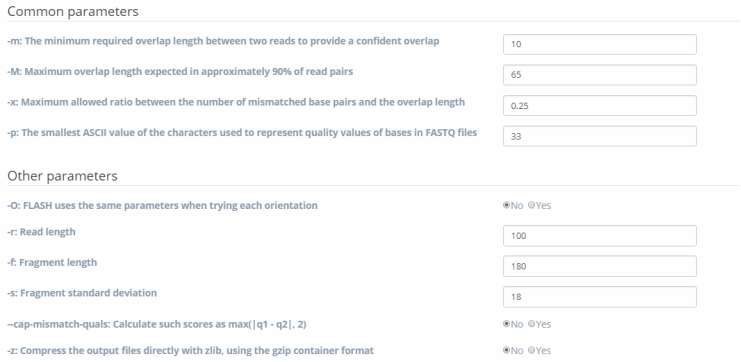 Figure 5. Uploading two files in FASTQ format.
Figure 5. Uploading two files in FASTQ format.Step 3: Then you can click the “Submit” button at the bottom of the interface to run the FLASH algorithm. Alternatively, users can also provide their email addresses in order to receive a notification Email after the submitted job is finished. Finally, when the submitted job is completed successfully, users can view the job details and download the generated results. In this case, users will receive a notification Email and can check the job details by clicking a hyperlink in the Email. The details are as shown in Figure 6.
 Figure 6. Submit job of FLASH algorithm.
Figure 6. Submit job of FLASH algorithm.Step 4: Get the result files and download them. After a few seconds, the interface will jump to the interface of download. The results with chosen descriptors are listed for download. The details are as shown in Figure 7.
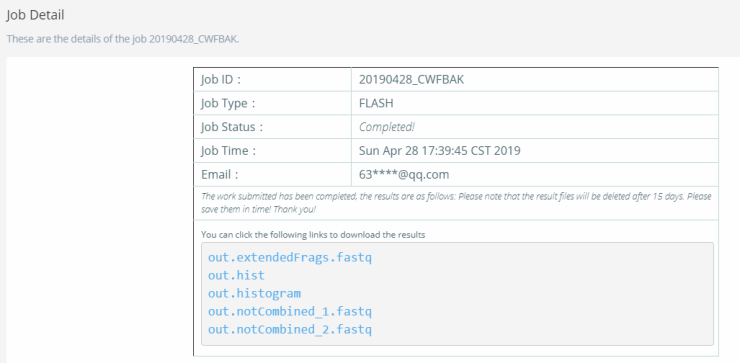 Figure 7. Download interface of FLASH algorithm.
Figure 7. Download interface of FLASH algorithm.2.2 Online PERF usage
The first three steps are similar to the usage of online FLASH. An illustration is shown in Figure 8-10. After users submit the job, the statistical table of the result page is shown Figure 11, which shows the results of PERF more directly. In the statistical table, users can input a favor SSR, then the statistical table only remains the rows with the favor SSR. In addition, users can download the complete result or the filtered result from this page.
 Figure 8. Input sequences with the FASTA format.
Figure 8. Input sequences with the FASTA format. Figure 9. Input parameter values of the PERF algorithm.
Figure 9. Input parameter values of the PERF algorithm. Figure 10. Submit job of the PERF algorithm.
Figure 10. Submit job of the PERF algorithm.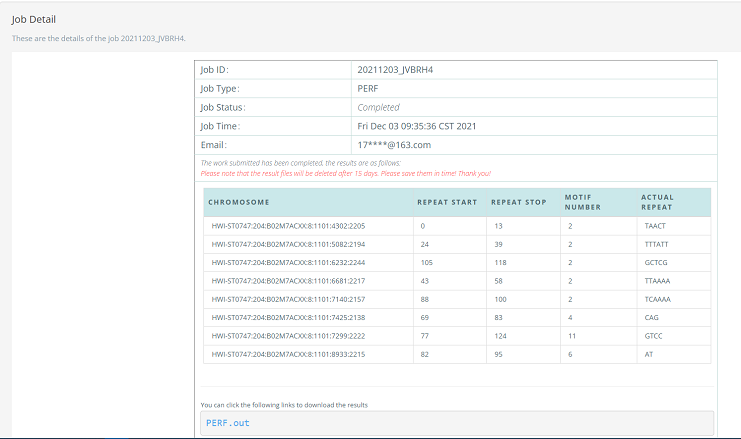 Figure 11. The statistical table of the result page of the PERF algorithm.
Figure 11. The statistical table of the result page of the PERF algorithm.2.3 Pipeline
This module integrates the FLASH algorithm with PERF into a pipeline. Users upload paired-end reads in the first place, then pass the parameter values to the FLASH and PERF , and finally submit this job. It is especially important to note that a new parameter about length of terminal flanking sequences is added in the pipeline. The pipeline usage process is similar to the usage of FLASH and PERF
